| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
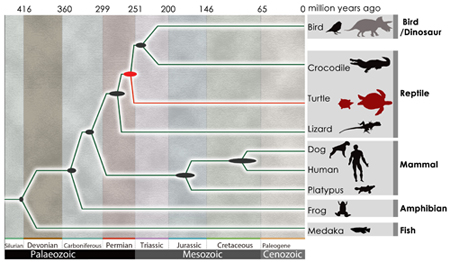
May 8, 2013 –The turtle is something of a monster of evolutionary developmental biology, not by virtue of its stern physiognomy or powerful jaw, but for its utterly unique morphology. The turtle carapace, for example, is made of up ribs that have protruded through the animal’s back and fused together in a bony shell, while its scapulae have moved to the body interior. Turtles also lack temporal fenestrae, small holes found in the skulls of other reptilians, further muddying the picture of their phylogeny. A study of a pair of turtle genomes by Naoki Irie and others in the Laboratory for Morphological Evolution (Shigeru Kuratani, Group Director) now sheds new light on its evolutionary history, establishing turtles as a sister group of crocodilians and birds (collectively, Archosauria), both of which diverged from the more ancient lizards and snakes (Lepidosauria). Published in Nature Genetics, this work reflects the collaborative efforts of an international turtle genome sequencing consortium with scientists from 11 institutions, including the Beijing Genomics Institute and Wellcome Trust Sanger Institute.
A number of competing hypotheses have been proposed to find the turtles’ true branch on the tree of life. These including placing turtles among early reptiles (anapsids), on account of their missing fenestrae; grouping them together with the snakes and lizards; and, categorizing them as archosaurs, along with crocodilians and birds. Using a massively parallel shotgun sequencing approach, the consortium generated and analyzed the genomes of the soft-shell turtle (Pelodiscus sinensis) and the green sea turtle (Chelonia mydas). Both genomes weighed in at about 2.2 billion base pairs, encoding around 19,000 genes. Comparisons with 10 other vertebrate genomes (including those of the zebrafish, frog, alligator, chicken and human) identified 1,113 orthologous genes, phylogenetic analysis of which revealed that turtles are most closely allied with the archosaurian birds and crocs. Using this data supplemented, the authors place the turtle divergence to around 268–248 million years ago. This conclusion agrees nicely with the fossil record, as the oldest known turtle dates back 220 million years. The apparent timing of their emergence, which would have coincided with the transitional period known as the Permian–Triassic Transition, characterized by large-scale extinctions, raises intriguing questions about possible linkage between the two events. Looking next to uncover details of the turtle genome itself, Irie et al. found an unexcpectedly large number of olfactory receptor genes, as many as 1,137 in the soft-shell turtle, a number similar to that possessed by mammals. Of these, many coded for group α olfactory receptors, used to detect hydrophilic odorant molecules, which may indicate that turtles have an excellent ability to sniff out variety of smells. A number of genes involved in taste, appetite, and metabolic regulation, however, were missing, which may reflect these animlas’ low-metabolic strategy. Previous work by Irie had focused on the developmental hourglass model that explains the evolutionary diversity of vertebrate embryos, and found support for the “hourglass” model, that in which homology is highest at a midpoint in development (called the vertebrate phylotypic period, during which the basic body plan is being established), and differs more widely before and after. To confirm whether turtles would also fit this scheme, the group used RNA sequencing to compare turtle and chicken, and found hourglass-like divergence in the embryogenesis of these taxa, indicating that molecularly conserved turtle embryos exhibit a common vertebrate developmental plan, after which turtle-specific patterning is likey to begin in earnest. The group not only identified 233 genes that showed turtle-specific expression patterns after the phylotypic stage, but also found the Wnt5a gene expressed both in the limbs and the carapacial ridge, suggesting possible involvement of Wnt signaling in carapace evolution. “Species make wide use of a shared toolbox of developmental processes and genetic information, but also generate new morphologies that could truly be called inventions. These new findings should be useful in guiding predictions about the course of vertebrate evolution,” says Kuratani. “But while these genomes are now sequenced, knowing the order of base pairs is not much more than being able to scan lines of text on a page. From now, we will need to explore how these genes are actually used during development to build body plans if we hope to decipher their true meaning.” *A gene involved in appetite was discovered in a subsequent analysis. [ref1, ref2] (Revised June 6, 2014) |
|||||
|
|||||
|
|||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |